About¶
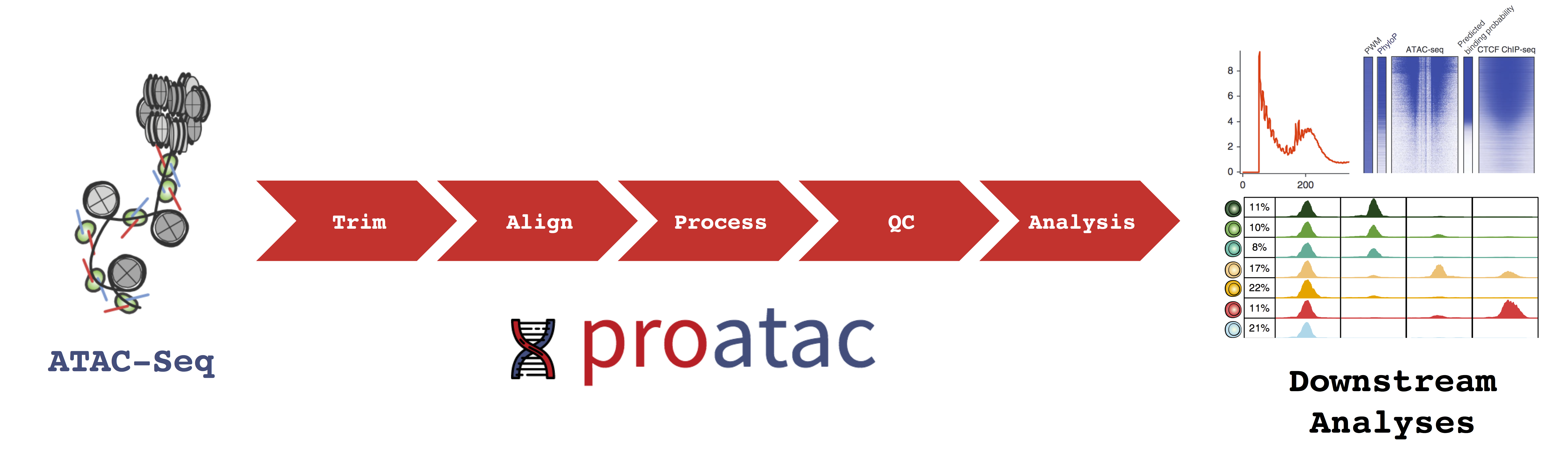
proatac is an open-source command-line toolkit that performs robust and scalable preprocessing of ATAC-Seq data. Specifically, we’ve implemented our workflow using Snakemake, a robust, scalable computational workflow platform. Various snakefiles are wrapped alongside meta-data annotations and various object-oriented constructs and distributed as a Python 3 package. The figure below provides a brief overview of the functionality of the proatac pipeline.
Installation¶
Dependencies¶
Trimming¶
Alignment¶
Annotating peaks¶
Author¶
The primary developer is Caleb Lareau in the Buenrostro Lab.
Citation¶
If you use proatac in your research, please cite our tool at the following URL:
http://buenrostrolab.com/proatac
Bugs / Errors¶
Please let us know if you find any errors/inconsistencies in the documentation or code by filing a new GitHub Issue.

